Department of Natural Products
Chemistry
Graduate School
of Pharmaceutical Sciences and
Plant Molecular Science Center
Chiba University
1-8-1 Inohana, Chuo-ku, Chiba 260-8675, JAPAN
Prof.
Masami ISHIBASHI, Ph.D.
@@TEL/FAX: +81-43-226-2923 @@
e-mail: mishchiba-u.jp gh should be replaced by g@h
Assoc.
Prof. Akiko TAKAYA, Ph.D.
@@TEL: +81-43-226-2924 @@
e-mail: akikofaculty.chiba-u.jp gh should be replaced by g@h
Assist.
Prof. Yasumasa Hara, Ph.D.
@@TEL: +81-43-226-2925 @@
e-mail: yharachiba-u.jp gh should be replaced by g@h
Comprehensive
Studies on Natural Products Chemistry: Chemical Biology based on
Natural Resources
Natural products continue to play an
important role in the discovery of low-molecular weight lead compounds for
new-drug developments. Research
interests of our laboratory are all concerned with chemistry of natural
products, mainly based on the search for new naturally occurring molecules from
a variety of terrestrial resources. Although extensive studies have been made
on isolation and identification of bioactive substances from plants and
microorganisms for more than a century, natural products chemistry is still of
great importance as a basic science since natural products have made
significant contributions to development of new drugs as well as progress of
basic studies of life sciences.
Research projects of our group are
aiming at (i) discovering bioactive small molecules useful for development of
new drugs and (ii) providing new molecular tools applicable to basic biological
sciences. These research projects
have to be carried out on the basis of modern organic chemistry, and advanced
skills are essentially required in isolation and structure elucidation of
natural organic compounds using current chromatographic techniques,
spectroscopic analyses, and chemical syntheses of key molecules.
Our current research
interests:
1)
Basic
studies on development of unexplored organisms such as myxomycetes which are
expected to be useful as new natural-products resources.
2)
Search
for new molecules with significant biological functions and unprecedented
chemical structures from a variety of natural resources.
3)
Synthesis
of bioactive natural products and their modified or fragmentary compounds for
the purpose of rigid determination of fine stereochemical structures and
application to the analysis of the mechanism of actions.
4)
Search
for bioactive small molecules from natural products through several screening
systems targeting mainly cancer-related signaling molecules using cell-based
luciferase or fluorescent assays.
5)
Construction
of small molecule library based on natural products framework by using
diversity-oriented synthesis and solid phase synthesis.
6)
Development
of high-throughput analyzing system of protein-protein interaction with small
molecule based on target protein immobilizing microplate.
Publications: Original Articles (1998-)
A summary for recent
studies in our group:
During our studies on search for bioactive
natural products from unexplored natural resources targeting singaling
molecules, here we describe two subjects: 1) Isolation of natural products from
myxomycetes; and 2) Search
for bioactive natural products targeting signaling molecules in cancer-related
biological pathways.
1.
Natural products from myxomycetes:
The
Myxomycetes (true slime molds) are an unusual group of primitive organisms that
may be assigned to one of the lowest classes of eukaryotes. Spore germination experiments were
studied of hundreds of field-collected myxomycetes collected in Japan and
succeeded in laboratory culture of plasmodia of several myxomycetes in a
practical scale for natural products chemistry studies.@Pyrroloiminoquinones,
polyene yellow pigments, and a peptide lactone were isolated from cultured
plasmodia of myxomycetes, while new antimicrobial naphthoquinone pigments,
tyrosine-kinase inhibitory bisindole alkaloids, a cytotoxic triterpenoid
aldehyde lactone
with a reversal effect of drug resistance, a
cycloanthranilylproline with sensitizing effect of TRAIL-induced apoptosis
through activation of COX2, a dibenzofuran glycoside, and sterols with a
2,6-dioxabicyclo[2.2.2]octan-3-one ring system were also
isolated from field-collected fruit bodies of myxomycetes (Figure 1).1-3)

Figure
1 New natural products isolated from
myxomycetes in our group
2. Search for bioactive
natural products targeting signaling molecules in cancer-related biological
pathways:
During our studies on
search for bioactive natural products, we recently examined extracts of various
natural resources including unexplored myxomycetes,1-3) marine
organisms,4) as well as several medicinal plants collected in
north-east part (Khon Kaen area) of Thailands.5) Here we describe our recent results on
our screening programs targeting signaling molecules in cancer-related
biological pathways such as TRAIL, Wnt, and Hedghog signaling pathways.
(1)
TRAIL signaling
Tumor necrosis factor
(TNF)–related
apoptosis-inducing ligand (TRAIL) induces apoptosis in many
transformed cells but not in normal cells and, hence, has been
expected as a new anticancer strategy. We recently identified several natural
products which exhibited activities related to TRAIL signaling (Figure 2).6) A dimeric sesquiterpenoid, parviflorene
F (1),7) isolated from Zingiberaceous plant, Curcuma
parviflora, showed enhancement activity of gene expression of
TRAIL-receptor (TRAIL-R2) and TRAIL-R2 protein level (Figure 3). Apoptosis was
induced by 1 as revealed by the distribution of DNA and
Annexin V/PI staining using flow cytometry. In addition, 1-induced
apoptosis was inhibited by human recombinant TRAIL-R2/Fc chimera protein,
TRAIL-neutralizing fusion protein.
We also found that 1 induced the activation of
caspase-8, caspase-9, and caspase-3, indicating that the cytotoxic effect of 1
is correlated with apoptosis by a caspase-dependent mechanism through
TRAIL-R2. In addition, 1
enhanced TRAIL-induced cell death against HeLa and TRAIL-resistant DLD1 cells.
Thus, it was suggested that up-regulation of TRAIL-R2 by 1 may
contribute to sensitization of TRAIL-induced cell death.8)
Several new isoflavone
natural products, named brandisianins (e.g., brandisianin D (2)),9)
were isolated from Leguminosaeous plant, Millettia brandisiana, by our screening
study targeting TRAIL receptor expression enhancement activity by a luciferase
assay system using DLD-1/SacI
cells. A dihydroflavonol (BB-1, 3)10)
that was extracted from
Compositaeous plant,
Blumea balsamifera, and fuligocandin B (4),11) a new
anthranilylproline-indole alkaloid isolated from myxomycete were found to
exhibit reversal effect of TRAIL resistance activity.
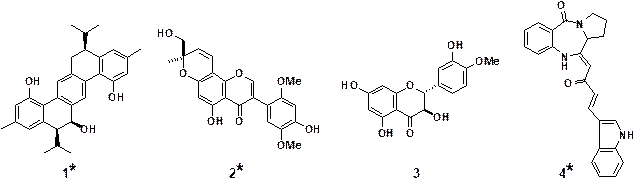
Figure 2. Natural products having
effects on TRAIL signaling
(*New compounds isolated
in our group)
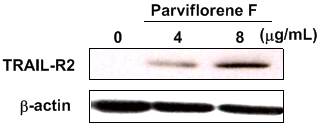
Figure 3. Enhancement of TRAIL-R2
protein levels in DLD1/TRAIL-R cells treated with parviflorene F (1)
at 4 and 8 Êg/mL (n = 3).
(2)
Wnt signaling
The Wnt/À-catenin signaling pathway plays
key roles in cell morphology, motility, proliferation, and differentiation. When inappropriately activated, the pathway has been linked to
colorectal cancer and melanoma. In these cells, the presence of Wnts or mutations
in APC, Axin, etc. cause activation of Wnt signaling and result in the
stablization of À-catenin. Then À-catenin enters into the nucleus and associates with transcriptional
factors of TCF/LEF, then this complex binds to the TCF/LEF binding sites and leads to the overexpression of target genes and finally
contributes to tumorgenesis. Therefore, compounds down-regulating Wnt/À-catenin signaling were expected in colon cancer therapy. To investigate small molecules down-regulating Wnt/À-catenin signaling,
natural extracts and compounds were tested using stably transfected cells (STF cells), in which luciferase reporter plasmids with TCF/LEF
binding sites were transfected. We
screened extracts of plants collected from Thailand, and five of them were judged as active.12) We also screened natural compounds from
myxomycetes isolated or synthesized by our laboratory.
Lycogarubin B (5), cis-dihydroarcyriarubin
C (6, prepared by synthesis),13,14) and 10-epi-melleumin
B (7, prepared by synthesis)15,16) were found to exhibit significant inhibition in TCF/À-catenin transcriptional
activity
(Figures 4 and 5).
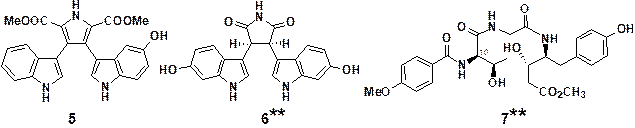
Figure 4. Natural products having
inhibitory effects on TCF/À-catenin
transcriptional activity
(**Stereoisomers of new
compounds isolated in our group and prepared by synthesis)
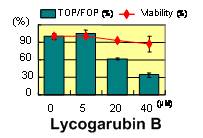
Figure 5. Inhibition of TCF/À-catenin transcriptional activity of lycogarubin B (5)
(3)
Hedghog signaling
The
hedgehog (Hh)/GLI signaling pathway has been implicated not only in a variety
of developmental processes in wide range of organisms, but also in the
formation and development of different tumors including skin, brain, prostate,
upper gastrointestinal tract, pancreas and lung. Targeting Hh/GLI signaling has been
expected as an effective cancer therapeutic strategy. To find specific inhibitors of Hh/GLI
signaling pathway from natural resources, a cell-based screening assay system
targeting transcriptional activator GLI1, which constitutes the final step in
the Hh signaling pathway, was constructed.
A pGL4-Luc reporter vector inserted with 12LGLI
binding sites was stably transfected into HaCaT cell line expressing GLI1 under
tetracycline repressor control.
By using this assay system, we identified six active compounds;
staurosporinone (8),
6-hydroxystaurosporinone (9),17)
arcyriaflavin C (10),
5,6-dihydroxyarcyriaflavin A (11),17)
zerumbone (12), and zerumbone
epoxide (13). Their IC50 values of GLI1
transcriptional inhibitory activity were 1.8, 3.6, 11.3, 6.9, 3.0 and 55 ÊM,
respectively. Next, from the screening study of our natural plants extracts
library, the extract of Physalis minima
was found to be active. Repeated chromatography separations of the MeOH
extracts of P. minima gave two active
compounds, physalin F (14) and
physalin B (15) with the IC50
values of 0.66 and 0.62 ÊM, respectively.
These
compounds also inhibited GLI2-mediated transactivation. These inhibitors are the first natural
products shown to be selective inhibition of GLI-mediated transcription. Western blotting analysis further
revealed that 8, 12, 14, and 15 decreased the
expression of GLI1 and PTCH proteins in HaCaT cells. It was also revealed for the first time
that these selective GLI-mediated transactivation inhibitors directly reduced
the level of the anti-apoptosis Bcl2 protein. Finally, physalins F and B were found to be cytotoxic against PANC1 pancreatic
cancer cells (IC50 values, 2.7 and 5.3 mM, respectively),
which significantly express Hh/GLI components. These results strongly suggested that
the cytotoxicity of these compounds against PANC1 cells may be correlated with
their inhibition of GLI-mediated transcription (Figures 6 and 7).18,19)
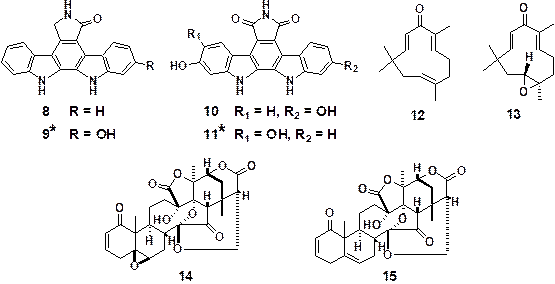
Figure 6. Natural products having
inhibitory effects on GLI transcriptional activity
(*New compounds isolated
in our group)
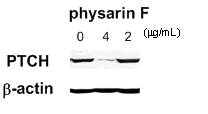
Figure 7. Decrease
in protein levels of PTCH, a Hh/GLI signaling component, in PACN1 cells treated
with physarin F (14)
References
and Notes
1) Ishibashi,
M. Yakugakuzasshi 2007, 127, 1369-1381.
2)
Ishibashi, M. Medicinal Chemistry 2005, 1,
575-590
3) Ishibashi, M.
Studies in Natural Products Chemistry, Atta-ur-Rahman, Ed.; Elsevier
Science; Amsterdam, 2003, 29, 223-262.
4) Ishibashi,
M.; Yamaguchi, Y.; Hirano, Y. J. Biomaterials from Aquatic and Terrestrial
Organisms, M. Fingerman and R. Nagabhushanam, Eds.; Science Publishers,
Inc.; Enfield, 2006, pp. 513-535.
5) Ishibashi,
M.; Toume, K.; Yamaguchi, Y.; Ohtsuki, T. Recent Research Developments in
Phytochemistry; Research Signpost; Trivandrum, India, 2004, 8,
139-156.
6) Ishibashi,
M.; Ohtsuki, T. Med. Res. Rev. 2008, 28, 688-714
7) Toume, K.; Takahashi, M.; Koyano, T.; Kowithayakorn,
T.; Yamaguchi,
K.; Hayashi, M.; Komiyama, K.; Ishibashi, M. Tetrahedron
2004, 60, 10817-10824.
8) Ohtsuki,
T.;
Tamaki, M.; Toume, K.; Ishibashi, M. Bioorg. Med. Chem. 2008, 16,
1756-1763.
9) Kikuchi, H.; Ohtsuki, T.; Koyano, T.; Kowithayakorn,
T.; Sakai, T.; Ishibashi, M. J. Nat. Prod. 2007,
70, 1910-1914.
10) (a)
Osaki, N.; Koyano, T.; Kowithayakorn, T.; Hayashi, M.; Komiyama, K.;
Ishibashi, M. J. Nat. Prod. 2005, 68, 447-449. (b) Hasegawa, H.; Yamada, Y.; Komiyama, K.; Hayashi, M.;
Ishibashi, M.; Yoshida, T.; Sakai, T.; Koyano, T.; Kam, T.-S.; Murata K.;
Sugahara, K.; Tsuruda, K.; Akamatsu, N.; Tsukasaki, K.; Masuda, M.; Takasu, N.;
Kamihira S. Blood
2006, 107, 679-688.
11) (a)
Nakatani, S.; Yamamoto, Y.; Hayashi, M.; Komiyama, K.; Ishibashi, M. Chem.
Pharm. Bull. 2004, 52, 368-370. (b) Hasegawa, H.; Yamada, Y.; Komiyama, K.;
Hayashi, M.; Ishibashi, M.; Sunazuka, T.;
Izuhara, T.; Sugahara, K.; Tsuruda, K.; Masuda, M.; Takasu, N.; Tsukasaki, K.;
Tomonaga, M.; Kamihira, S. Blood 2007, 110,
1664-1674
12) Li,
X.; Ohtsuki, T.; Koyano, T.; Kowithayakorn,
T.; Ishibashi, M. Chem. Asian J.
2009,
4, 540-547
13) Kaniwa,
K.; Arai, M. A.; Li, X.; Ishibashi, M. Bioorg. Med. Chem. Lett. 2007,
17, 4254-4257.
14) Nakatani,
S.; Naoe, A.; Yamamoto, Y.; Yamauchi, T.; Yamaguchi, N.; Ishibashi, M. Bioorg.
Med. Chem. Lett. 2003, 13, 2879-2881.
15) Hanazawa, S.; Arai, M. A.; Li, X.; Ishibashi, M.
Bioorg. Med. Chem. Lett. 2008, 18,
95-98.
16) Nakatani, S.; Kamata, K.; Sato, M.; Onuki,
H.; Hirota, H.; Matsumoto, J.; Ishibashi, M. Tetrahedron
Lett. 2005, 46, 267-271.
17) Hosoya,
T.; Yamamoto, Y.; Uehara, Y.; Hayashi, M.; Komiyama, K.; Ishibashi,
M. Bioorg. Med. Chem. Lett. 2005, 15, 2776-2780.
18) (a) Hosoya, T.; Arai, M. A.; Koyano, T.;
Kowithayakorn, T.; Ishibashi, M. ChemBioChem 2008,
9, 1082-1092. (b) Arai, M. A.;
Tateno, C.; Hosoya, T.; Koyano, T.; Kowithayakorn, T.; Ishibashi, M. Bioorg.
Med. Chem. 2008, 16, 9420-9424
19) Acknowledgment:
Myxomycetes are collected by Yukinori
Yamamoto (Ohtsu-ko, Kochi), and plant materials are provided through a
collaboration project with Dr. Takashi Koyano (Temko Corporation) and Professor
Thaworn Kowithayakorn (Khon Kaen Univeristy, Thailand) or a collaboration
project with Professor S. K. Sadhu (Khulna University, Bangladesh). We thank
Dr. Bingliang Fang (The University of Texas, MD, Anderson Cancer Center) for
TRAIL-resistant DLD1 cells, Prof. T. Sakai (Kyoto Prefectural University of
Medicine) for DLD-1/SacI cells, Prof. J. Nathans (John Hopkins Medical
School) for the STF cells, Prof. F. Aberger (University of Salzburg) for tetracycline-regulated
HaCaT cells, and Prof. R. Toftgård (Karolinska Institute) for the 12GLI-RE-TKO
luciferase plasmid. We also thank Dr. Masaaki Sato for
valuable discussions in the beginning of these studies. This work was supported by a
Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and
Technology (MEXT) of Japan, from the Futaba Electronics Memorial Foundation and
the
Japan Science and Technology Agency.